Usage
To use dicompyler-core in a project:
DICOM data can be easily accessed using convenience functions using the dicompylercore.dicomparser.DicomParser class:
from dicompylercore import dicomparser, dvh, dvhcalc
dp = dicomparser.DicomParser("rtss.dcm")
# i.e. Get a dict of structure information
structures = dp.GetStructures()
>>> structures[5]
{'color': array([255, 128, 0]), 'type': 'ORGAN', 'id': 5, 'empty': False, 'name': 'Heart'}
Dose volume histogram (DVH) data can be accessed in a Pythonic manner using the dicompylercore.dvh.DVH class:
rtdose = dicomparser.DicomParser("rtdose.dcm")
heartdvh = dvh.DVH.from_dicom_dvh(rtdose.ds, 5)
>>> heartdvh.describe()
Structure: Heart
-----
DVH Type: cumulative, abs dose: Gy, abs volume: cm3
Volume: 437.46 cm3
Max Dose: 3.10 Gy
Min Dose: 0.02 Gy
Mean Dose: 0.64 Gy
D100: 0.00 Gy
D98: 0.03 Gy
D95: 0.03 Gy
D2cc: 2.93 Gy
>>> heartdvh.max, heartdvh.min, heartdvh.D2cc
(3.0999999999999779, 0.02, dvh.DVHValue(2.9299999999999815, 'Gy'))
Dose volume histograms (DVHs) can be independently calculated using the dicompylercore.dvhcalc module:
# Calculate a DVH from DICOM RT data
calcdvh = dvhcalc.get_dvh("rtss.dcm", "rtdose.dcm", 5)
>>> calcdvh.max, calcdvh.min, calcdvh.D2cc
(3.0899999999999999, 0.029999999999999999, dvh.DVHValue(2.96, 'Gy'))
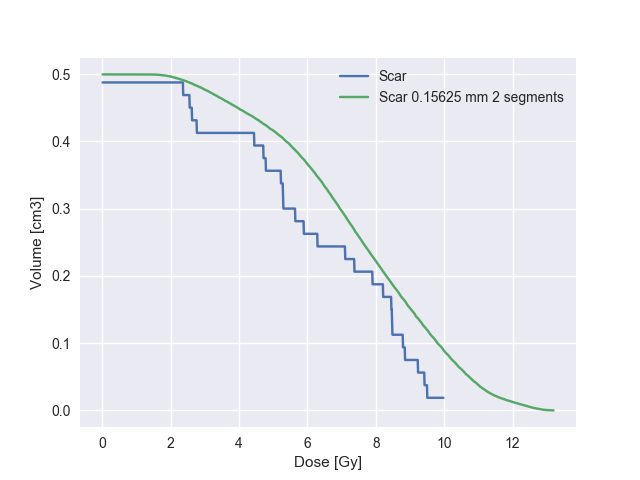
Structure and dose data for independently calculated DVHs can also be interpolated in-plane and between planes:
# Calculate a DVH using interpolation to super-sample the dose grid in plane,
# interpolate dose between planes and restrict calculation to the structure
# extents
interpscar = dvhcalc.get_dvh("rtss.dcm", "rtdose.dcm", 8,
interpolation_resolution=(2.5/16),
interpolation_segments_between_planes=2,
use_structure_extents=True)
interpscar.name += ' interp'
# Compare against un-interpolated DVH
origscar = dvhcalc.get_dvh("rtss.dcm", "rtdose.dcm", 8)
>>> origscar.compare(interpscar)
Structure: Scar Scar interp Rel Diff Abs diff
-----
DVH Type: cumulative, abs dose: Gy, abs volume: cm3
Volume: 0.47 cm3 0.50 cm3 +6.55 % +0.03
Max: 9.50 Gy 13.18 Gy +38.74 % +3.68
Min: 2.36 Gy 1.23 Gy -47.88 % -1.13
Mean: 6.38 Gy 7.53 Gy +18.02 % +1.15
D100: 0.00 Gy 0.00 Gy +0.00 % +0.00
D98: 2.36 Gy 2.44 Gy +3.39 % +0.08
D95: 2.36 Gy 3.09 Gy +30.93 % +0.73
D2cc: 0.00 Gy 0.00 Gy +0.00 % +0.00
Dose grids can be summed and scaled using the dicompylercore.dose module:
from dicompylercore import dose
# Dose grid summation with (tri-linear) interpolation if dose grids are not spatially coincident
grid_1 = dose.DoseGrid(dose_file_1)
grid_2 = dose.DoseGrid(dose_file_2)
grid_sum = grid_1 + grid_2
grid_sum.save_dcm("grid_sum.dcm") # save to file
# Dose grid scaling
grid_scaled = grid_1 * 2 # Scale grid_1 by a factor of 2
grid_scaled.save_dcm("grid_scaled.dcm") # save to file
# Dose grid subtraction may be performed, however, negative doses are not currently
# DICOM compliant (i.e., the pixel_array of RTDOSE datasets are unsigned integer arrays).
# dicompyler-core users must work with the DoseGrid's numpy array directly (DoseGrid.dose_grid)
dose_diff_direct = grid_1.dose_grid - grid_2.dose_grid
dose_diff_interp = grid_1.dose_grid - grid_1.interp_entire_grid(grid_2)